Science coding vs. Software development
My opinion on the differences between science coding and software development and how to mix the two.
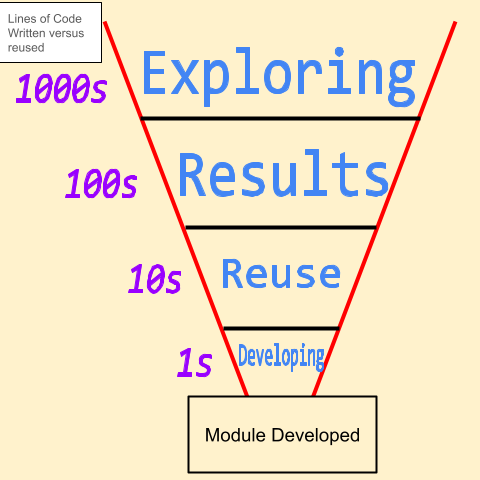
I have this idea that the number of lines of code written in the name of science follows a power law distribution. For every 1000 lines of code written exploring data, 100 may turn into a result, 10 may be re-used at any time in the future, and 1 of those lines of code end up in a module somewhere (see the fancy plot I made below). This is in contrast to developing software where the exploration phase may be done in meetings with colleagues trying to design a piece of software to solve a particular problem. And while I recognize that software dev teams often develop using agile coding practices, in my experience, software developers have more of a gaussian relationship between lines of code and utility. This is because design specifications simply change less frequently than data exploration.
I think the reason software specs change less frequently than data exploration is because there is an inherent difference in the goal of the scientist to the software developer. The scientist has a set of data that initially is diffuse and poorly understood. As they try out different ways of transforming and interacting with the data, they begin to find things in that data that was otherwise not apparent from the start. These data transformations are typically custom enough to the data set that it is easier to write a block of code to do the transformation and see if it shows something instead of developing a module that does something either as one line of code or as an application.
For example, transforming some kind of signal into information largely requires you to examine the signal both in amplitude space and in frequency space. In order to do this you use the fourier transform to calculate the frequency space. Windowed over time this produces a spectrogram. However there are all sorts of bits and pieces to consider when making such a calculation. What is the sample rate of the data? Do you want to do some kind of filtering prior to plotting a spectrogram? What time window size do you want? Do you want to focus only a subset of the frequency range or the entire range? Do you want to do this 1 million times to generate spectrograms to use for some kind of image-based machine learning signal finder? Ultimately, it ends up being easier to use a scripting language to do this instead of an application with knobs and buttons. But all this code may likely never be used again. Thus, it all falls into the “Exploring” and “Results” category above.
This conceptual relationship between how many lines of code you write versus how many of those lines of codes get reused ends up being a big sticking point and why you end of with questions about why are scientists so bad at writing code. Answers usually include that scientists have a different purpose than software devs. But what if you do want to build software for science? How do you get buy-in from highly technical scientists who may have poor software development skills? How do you create a conversation between the scientists and the software devs so everyone is on the same page? Enter tutorial driven software development.
Tutorial driven software development is a software development paradigm being developed by the brilliant Christopher Woods who formerly headed up the Research Software Engineering group at University of Bristol and now is heading up AI/supercomputing infrastructure group. Tutorial driven software development positions the scientific problem first by developing tutorials for using the software prior to the software library being written. This allows the software developers to sit with the scientists and work together to generate a set of tutorials that solves the problem of the scientist with software. This is done in a cycle. First you develop a draft of a tutorial. This tutorial is typically done via a jupyter notebook which allows scientists to write content and context via markdown along with function stubs that implement the library. These function stubs serve as the endpoints of the software library to be developed. The development is then pursued by software devs, and this is iterated on until the tutorial is able to be leveraged by the scientists and extended into their work. The tutorials also serve as the ideal documentation that faces scientists who are more likely to search for documentation that details their specific use cases via tutorials, than read documentation at the library level.
Christopher has used this software development paradigm to develop multiple packages including the 4d-modeller R package that we just wrapped up development at the end of last year. I really appreciate that a core part of this development paradigm respects the different coding and computational skills across a diverse group. It allows domain expert stakeholders to be an integral part of the development process leveraging their expertise while also respecting that they may not have the time nor background to develop a sophisticated piece of software as they have other project responsibilities.